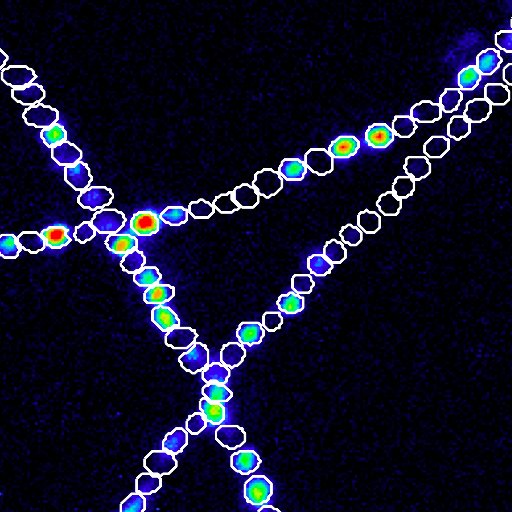
The #expobiome is comprised of #microbiome-derived #DNA, #RNA, #proteins & #metabolites can trigger many human pathways in disease. 2/8
The #expobiome, comprised of #microbiome-derived #DNA, #RNA, #proteins & #metabolites, can trigger many human pathways in disease. 2/8
@lcsb_uni_lu @uni_lu @velma_te_a The excitement continues with @mara_lucchetti with the talk “Emulating the gut-liver axis: Dissecting the microbiome’s effect on drug metabolism using multi-organ-on-chip models” 💊🦠 at #ISME18 on Tuesday at 12 PM, room 1ABC, session MS11
Combining extremely highly resolved metagenomics w/ personalized in vitro models, e.g., our biomimetic gut-on-chip model #HuMiX (https://t.co/3arm5tmrja), will lead to novel insights into individual-resolved host-microbiome crosstalk to better understand human health and disease
Brilliant artwork by @wampachl summarising our baby-#microbiome studies so far. Check out the latest #COSMIC3 paper 👇. @uni_lu @ULResearch @CHL_Luxembourg @microh_dtu @ResearchLux @FnrLux @ERC_Research https://t.co/g9jwiCJJO2
Grateful for the amazing contributions by @LauradeNies SaraLopes @_A_H_B_ @claczny @susheelbusi @GalataValentina PatrickMay @MicroSysEco @microh_dtu @uni_lu @ResearchLux. Funding by @FnrLux @ERC_Research. Art by @GalataValentina. 2/3
🏄the🌊of change. Great artwork by @GalataValentina about our recent paper in @NatureComms https://t.co/TbOtAk1f2S #ScienceMeetsArt #SciArt
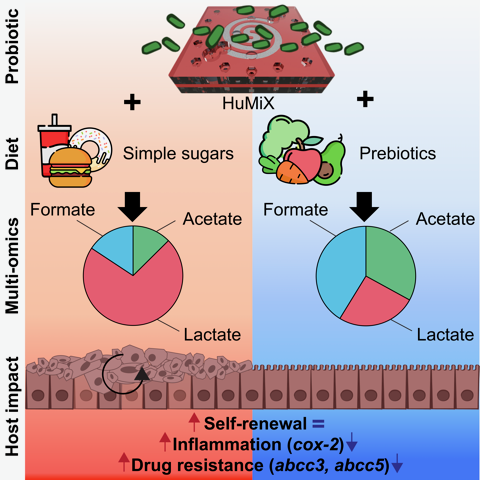
nutriHuMiX paves the way towards #personalized models for rationally combining #synbiotics with anti- #cancer #therapy in future @uni_lu @FnrLux @CellReports #openaccess https://t.co/Cm4lwmlmGB
nutriHuMiX out now in @CellReports - Developing our #OrganOnChip gut #microbiome model to study individual and combined effects of #diet and #probiotics on #cancer cells @uni_lu @FnrLux https://t.co/Cm4lwmlmGB