PDBのTwitterイラスト検索結果。 222 件中 6ページ目
This week: 509 SARS-CoV-2 structures freely available in the PDB archive https://t.co/C8psgBQVrw
#inktober2020 Day15 #Outpost
Toll-Like receptors, the brave sentinels of our immune system! They act in the first line of defence against bacteria and viruses, triggering an inflammatory fight against intruders
#inkthescience #scicomm #sciart #Immunology
inspired by PDB 3fxi
Dexamethasone and Cytokine Storms: Preventing too much of a good thing during SARS-CoV-2 infection
New feature in PDB-101's Resources to Fight the COVID-19 Pandemic https://t.co/vNStgOYtuW
Congratulations to PDB depositors Emmanuelle Charpentier and Jennifer A. Doudna on the Nobel award for the development of a method for genome editing.
Cascade and CRISPR help bacteria remember how to fight viral infection. More at Molecule of the Month https://t.co/39SeogATE3
6 new SARS-CoV-2 structures in the PDB this week (321 total). Access these structures and more at https://t.co/C8psgBQVrw
When will we reach 400?
A new representation of polymeric carbohydrates has been introduced in the PDB. This new representation includes information about carbohydrate linkages and standardised nomenclature. To find out more visit the wwPDB page at https://t.co/5CTanOH2yM 1/2
This week's PDB release has almost 300 new entries, including this O-methyltransferase structure from the strawberry, published in @ActaCrystF. View all the new PDB entries at https://t.co/54E03LeH5m
Changing character names on PDB until I get IP banned 💖 here’s the highlight reel ✨
There are 230 new #PDB entries this week, including this janus kinase structure, bound to the dermatological drug Delgocitinib, published in @JMedChem. View all the new PDB structures at https://t.co/54E03LeH5m
#Bornonthisday in 1943 was American chemist Richard Smalley. He shared the Nobel Prize in Chemistry with Harry Kroto and Robert Curl for their discovery of buckminsterfullerene or "buckyballs". There are 3 entries in the PDB with buckyballs! Find them at https://t.co/aNHm3pLY6x
Newly available trajectories for the SIRAH-CoV2 initiative:
S1 Receptor Binding Domain in complex with human antibody CR3022 (PDB 6W41) https://t.co/QqE6AdP0An
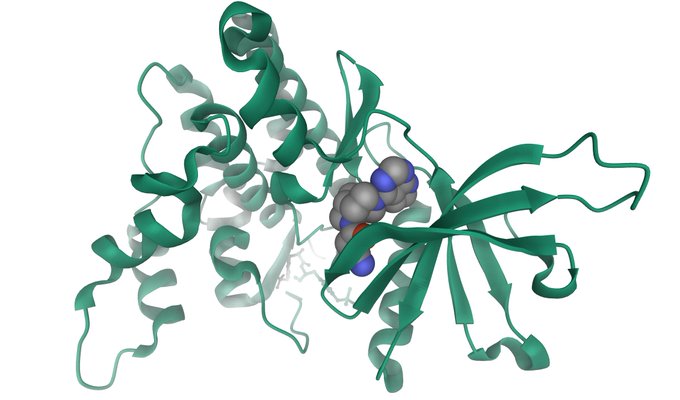
NSP16 - NSP10 Complex (PDB 6W4H) https://t.co/wrqF6pJDPX (image below)
#COVID19
This week's #PDB release sees another 16 structures of the #SARSCoV2 main protease with different fragments bound, solved by researchers at @DiamondLightSou. All this structural information and more is available at our #PDBeKB #COVID19 data portal: https://t.co/N6oiomu2ZC
Watercolor & ink painting of the COVID-19 main protease
#SciArt #SciArtSunday #coronavirus
-- based on @buildmodels PDB structure 6LU7, 2019-nCov coronavirus 3CL hydrolase (Mpro)
https://t.co/2fTwfVkcHa
New in this week's release is this #cryoEM structure of the #SARSCoV2 RNA polymerase (7btf). Remember, you can find all available PDB data on #COVID19, including ligand binding and interaction sites, at our PDBe-KB COVID-19 data portal: https://t.co/NBFmsXI9VW
This week's #PDB release includes 68 structures of #COVID19 protease, with various bound fragments identified using PanDDA analysis. Work by @MartinWalshDLS, @Darenfearon, @londonlab and more @diamondLightSou. Find all new #COVID19 structures at https://t.co/cUduRLl7OE
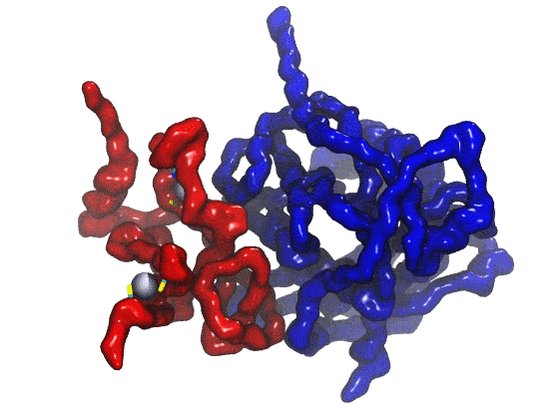
The earliest PDB example of ribonuclease was available in 1979 (now PDB structure 5rsa), but had been described in scientific literature previously. Deposition of structure data was not always a requirement for publication
Ribonuclease: https://t.co/QWHirE051A