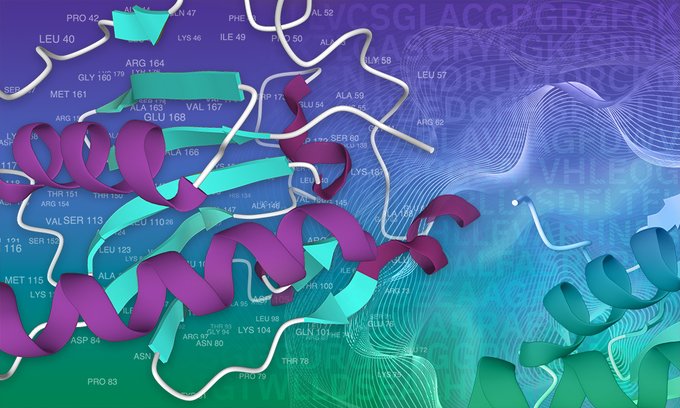
Thanks to #deeplearning models, @PfamDB – now accessible in @InterProDB – is continuing to expand.
Find out how this collaboration with @GoogleAI is increasing the number of protein annotations in the database.
https://t.co/wibsY5ABTx
AI methods can help scientists analyse #cryoEM data.
Find out how researchers from @UniHalle developed a new method to automatically interpret cryo-EM data using open source #AI software including #AlphaFold. #KastritisLab
https://t.co/gmuPFFkLhT
With help from #deeplearning models, @PfamDB has expanded by almost 10% exceeding all expansion made to the database over the last decade 🤯
@Alexbateman1 discusses this work and what the future holds for protein family classification.
https://t.co/wibsY5j11Z
This is just one of the many new initiatives under the Infection Biology theme of @embl's new scientific programme Molecules to Ecosystems, which we’re proud to be a part of #beatNTD #EMBLInfectionBiology
https://t.co/sMRmNIXc0m
#AI for protein structure prediction is the @ScienceMagazine breakthrough and a @Nature top pick of the year 🎉
Thousands of protein structure predictions powered by @Deepmind's revolutionary AI are available in the #openaccess AlphaFoldDB
https://t.co/yV1ebwvnlV
Introducing Milo: an #opensource R software package that generates high-resolution differential abundance testing results of cell states from your #singlecell data. Find out more and give Milo a try 👇 @MarioniLab @NatureBiotech @sangerinstitute @CRUK_CI
https://t.co/4mfURvTEq2
.@Ensembl 91 has been released, with a host of new and updated primate genomes and much more! https://t.co/1wKOM9w9TV
Happy #DNADay, everyone! #DNA is a common thread in our #dataservices, which are visited ~27 million times a day. https://t.co/OSUGta5vbs