alphafoldのTwitterイラスト検索結果。 31 件
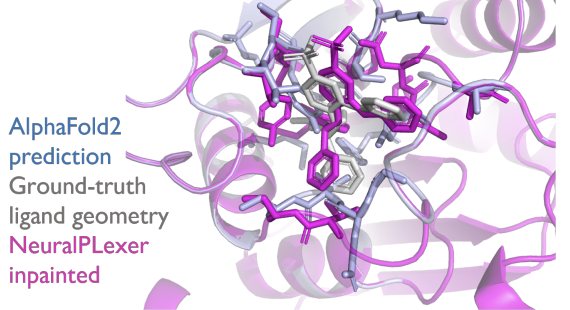
#Diffusion models are effective for protein-ligand structure prediction with equivariant transformers. Our method NeuralPLexer accurately inpaints binding site while using #alphafold doesn't. @ZhuoranQ @wn8_nie @ArashVahdat @tfmiller3 https://t.co/Yz1Wi2hTUs
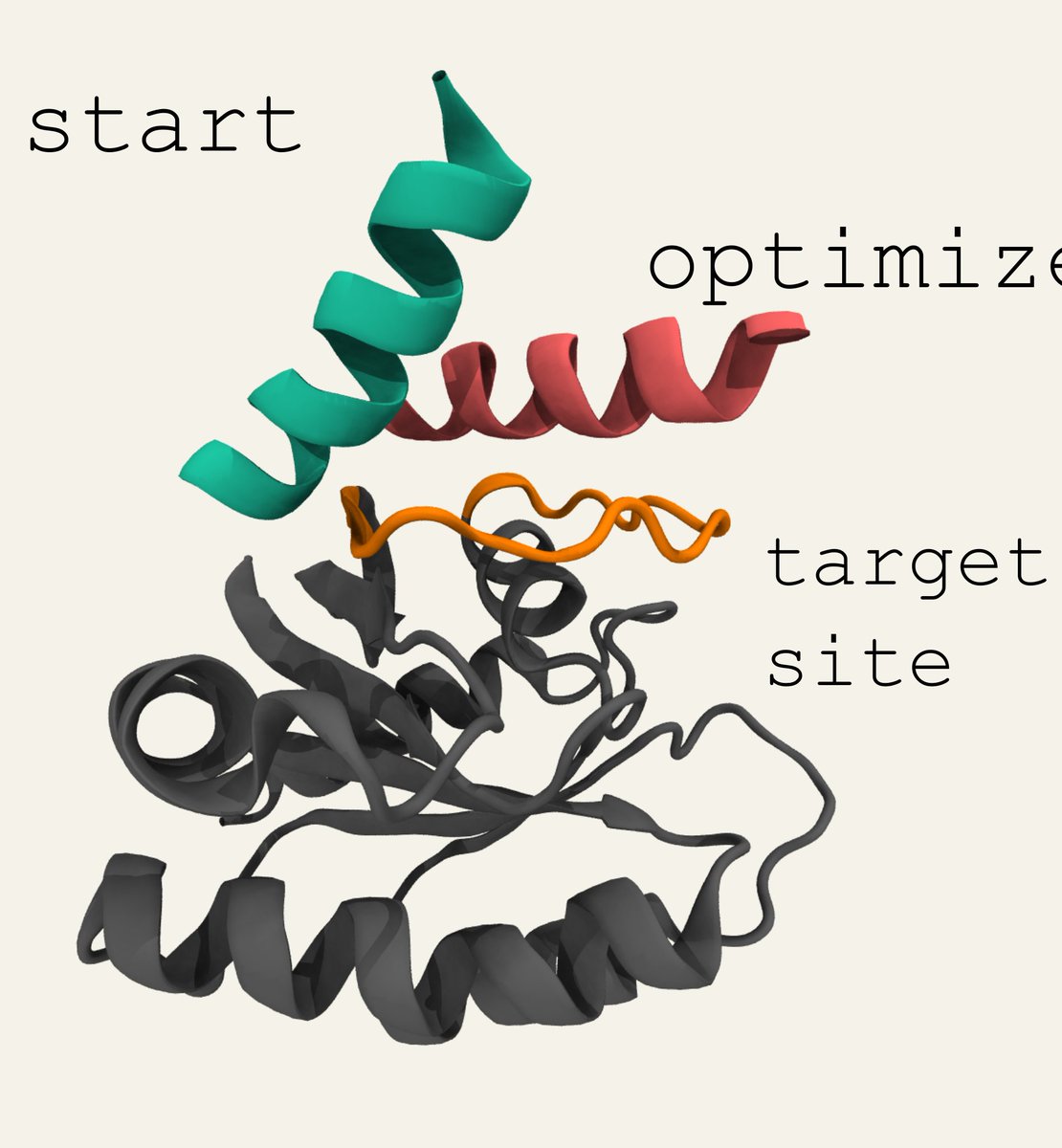
Packaged our algorithm to design binding peptides into a simple colab. Enter a protein sequence, indicate where you want the peptide to bind, and in a few minutes you'll have a peptide predicted to tightly bind via AlphaFold + Bayesian Optimization. 1/2
https://t.co/MvoXmnUHvK
AI methods can help scientists analyse #cryoEM data.
Find out how researchers from @UniHalle developed a new method to automatically interpret cryo-EM data using open source #AI software including #AlphaFold. #KastritisLab
https://t.co/gmuPFFkLhT
A new era for structural biology
"The AI revolution allows us to peer inside something that is not visible to us from the outset"
A. Ramanathan
#Science
#ArtificialIntelligence
#AlphaFold
⏯️Researchers turn to #deeplearning to decode protein structures
https://t.co/o8gmPsbXbq
Finally got round to drawing @DeepMind ‘s proteins folding predictor #AlphaFold as a man. #sciart
#AI for protein structure prediction is the @ScienceMagazine breakthrough and a @Nature top pick of the year 🎉
Thousands of protein structure predictions powered by @Deepmind's revolutionary AI are available in the #openaccess AlphaFoldDB
https://t.co/yV1ebwvnlV
Interesting... I suspect, by reducing the # of seqs in the MSA, this reduced the strength/certainty of restraints derived from coevolution, allowing AlphaFold to generate multiple hypotheses (conformations) with randomseed, model param, or msa pertubations https://t.co/3JPM8Ysgox
Recording of 'FEBS Junior Sections – AlphaFold Database webinar' by @PDBeurope team leaders - Gerard Kleywegt and Sameer Velankar is now available. Find it here https://t.co/2yCepK5YsD
#AlphaFold @FEBSnews
The #AlphaFold source code has been updated and now accounts for multi-chain protein complexes - providing a significant improvement in accuracy for predicting protein interactions: https://t.co/4gyZ0loLrd
Generate predictions from your browser via: https://t.co/2Vd4itjElu
A paper on "AlphaFold-multimer", a version of AlphaFold that works on protein complexes, was released by @DeepMind.
Accurately predicted structures can lead to better understand the function of such protein complexes that underpin many biological processes!
#DeepLearning 1/4
Many single protein chains can be accurately predicted using AlphaFold, but multi-chain protein complexes remain challenging. Introducing AlphaFold-Multimer, a model that can predict the structure of multi-chain protein complexes with increased accuracy https://t.co/KMyN87wgDE 1/
Spot the difference: can you find all 49359 differences between these two #AlphaFold galleries?
It takes only seconds to model glycans with GlycoSHIELD https://t.co/U87xBZMGiF.
It is indeed #glycotime!
Glad to see this out! Minor contribution from my side. We tested to see how well AlphaFold can predict homo-oligomers and their oligomeric state. Here I'm plotting the min(pTMscore) across 5 models, splitting success (top row) and failures (bottom row). https://t.co/fhdMtC3f4L
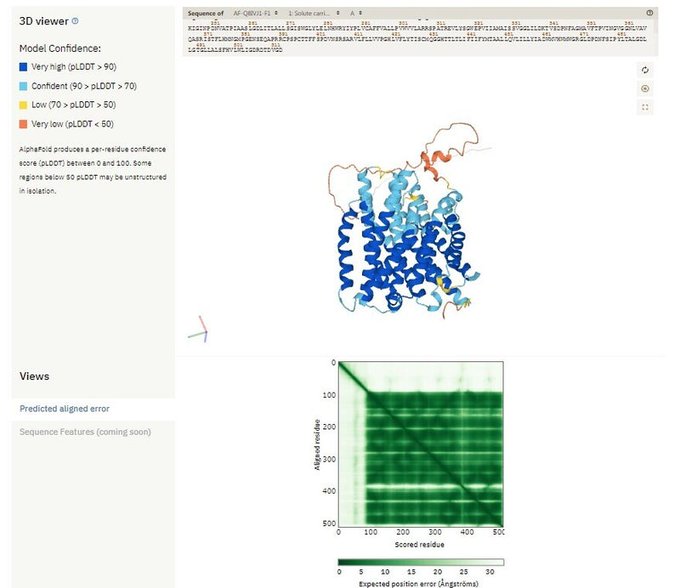
A #MachineLearning method, AlphaFold2, "predicts protein structures with near-experimental accuracy."
I read again with pleasure about this breakthrough!
#Science
#AI
@emblebi
⏯️Protein-structure prediction revolutionized
1-https://t.co/4r1fmqRXRc
2-https://t.co/LKXDQpJOGf
/AlphaFold2の非専門家向け活用法 第4回前半「AlphaFoldが現状カバーできていない構造情報」|上海老師|note https://t.co/GNnuOGbV6n
Like everybody else in the world of science, I've been having fun exploring the database of AlphaFold structures. Here are (subunits of) a few proteins that show up often in my paintings. It (amazingly) gets all the bits and pieces right, but doesn't quite yet get the biology.
分子収斂が見つかったら勝手にAlphaFoldの構造をダウンロードして収斂座位をマップしたPyMOLセッションを作ってくれる機能を実装してみた。
遅ればせながら、ようやく時間ができたのでAlphaFold2で遊んでる。ATeamで使ってる枯草菌のFoF1-εの配列入れて見たところ、リガンドのATP無くてもC末端ヘリックスバンドルが折りたたまれた結果が出てきた。これは予想外。